Genetically Defined Mouse Ovarian Cancer Cell Lines
- List of available cell lines Orsulic mouse ovarian cancer cell lines list (PDF link)
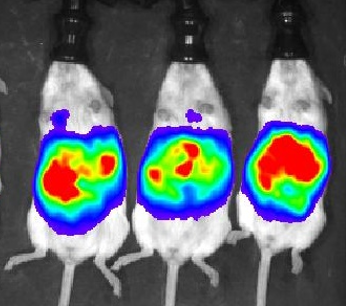
Orthotopic inoculation of syngeneic mouse ovarian cancer cell lines into FVB mice (PDF link) recapitulates key aspects of human ovarian cancer pathophysiology. These models are suitable for preclinical testing of drugs that may interfere with the immune system or testing the roles of the immune system in ovarian cancer progression.
RCAS plasmids (myr-Akt, myc, K-ras) can be obtained from Addgene http://www.addgene.org/pgvec1 (search Orsulic).